

| The story is this: I bought a car out of state, and I'm trying to get the safety inspection in Pennsylvania. The problem is that the car has aftermarket tint on all windows except the windshield. The tint is rather weak, and you can clearly see the inside of the car through the tint. The inspection garage said that they won't pass it unless I get a waiver from the state police. So I went to the state police - the officer told me that aftermarket tint is illegal, and I can get a waiver only for a pre-84 car or for a medical reason. I asked him to show me the section of the vehicle code that says it's illegal. He showed it to me and the paraghaph said that you can't have tint, if you can't see the inside of the car because of the tint. When I told him that you can in fact see the inside very well, he shut the book and said "It's just illegal, and in fact we can have someone give you a ticket for it right now." Well, won't argue with that... | I do not have enough medical expertise to have much of an opinion one way or another on hidden candida infections. I can understand the skepticism of those who see this associated with various general kinds of symptoms, while there is a lack of solid demonstration that this happens and causes such general symptoms. (To understand this skepticism, one only needs to know of past failures that shared these characteristics with the notion of hidden candida infection. There have been quite a few, and the proponents of all thought that the skeptics were overly skeptical.) On the other hand, I am happy to read that some people are sufficiently interested in this possibility, spurred by suggestive clinical experience, to research it further. The doubters may be surprised. (It has happened before.) |
| rec.autos | sci.med |

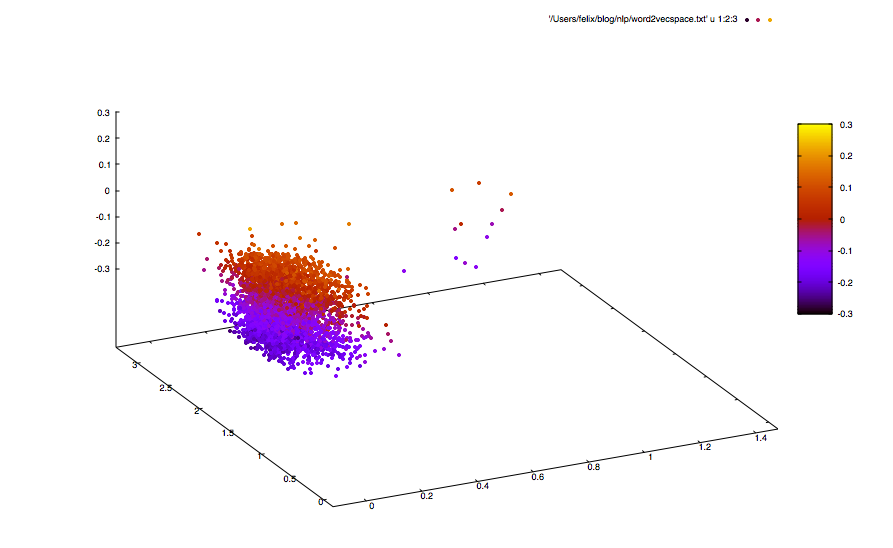
bin/word2vec -train all.txt -output all-vec.txt -debug 2 -size 20 -window 10 -sample 1e-4 -negative 5 -hs 0 -binary 0 -cbow 0
the -0.127186 0.277165 -0.027995 -0.057839 0.131760 -0.279101 -0.412328 -0.299498 0.064663 -0.325453 0.196605 ...
a -0.052591 0.278205 0.023183 -0.106372 0.146495 -0.280094 -0.375689 -0.279042 0.076930 -0.291453 0.167866 ...
to -0.071252 0.301636 0.012357 -0.089838 0.157550 -0.289630 -0.425848 -0.288347 0.048744 -0.313050 0.204709 ...
...
import neuroflow.application.plugin.IO._
import neuroflow.application.plugin.Style._
import neuroflow.application.processor.Util._
import neuroflow.core.Activator._
import neuroflow.core._
import neuroflow.dsl._
import neuroflow.nets.cpu.DenseNetwork._
val netFile = "/Users/felix/github/unversioned/ct.nf"
val maxSamples = 100
val dict = word2vec(getResourceFile("file/newsgroup/all-vec.txt"))
def readAll(dir: String, max: Int = maxSamples, offset: Int = 0) =
getResourceFiles(dir).drop(offset).take(max).map(scala.io.Source.fromFile)
.flatMap(bs => try { Some(strip(bs.mkString)) } catch { case _ => None })
def readSingle(file: String) = Seq(strip(scala.io.Source.fromFile(getResourceFile(file)).mkString))
def strip(s: String) = s.replaceAll("[^a-zA-Z ]+", "").toLowerCase
def normalize(xs: Seq[String]): Seq[Seq[String]] = xs.map(_.split(" ").distinct.toSeq)
def vectorize(xs: Seq[Seq[String]]) = xs.map(_.flatMap(dict.get)).map { v =>
val vs = v.reduce((l, r) => l.zip(r).map(l => l._1 + l._2))
val n = v.size.toDouble
vs.map(_ / n)
}
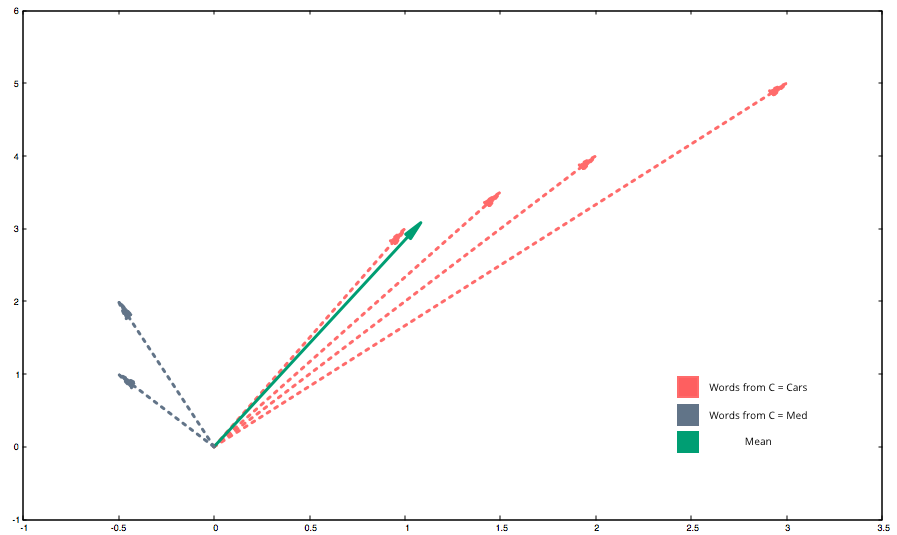
The mean word of a text, simplified in ℝ2
val cars = normalize(readAll("file/newsgroup/cars/"))
val med = normalize(readAll("file/newsgroup/med/"))
val trainCars = vectorize(cars).map((_, ->(1.0, 0.0)))
val trainMed = vectorize(med).map((_, ->(0.0, 1.0)))
val allTrain = trainCars ++ trainMed
println("No. of samples: " + allTrain.size)
val L =
Vector (20) ::
Dense (40, Tanh) ::
Dense (40, Tanh) ::
Dense (2, Sigmoid) :: SoftmaxLogEntropy()
val net = Network(
layout = L,
settings = Settings[Double](iterations = 15000, learningRate = { case _ => 1E-4 })
)
net.train(allTrain.map(_._1), allTrain.map(_._2))
File.write(net, netFile)
_ __ ________
/ | / /__ __ ___________ / ____/ /___ _ __
/ |/ / _ \/ / / / ___/ __ \/ /_ / / __ \ | /| / /
/ /| / __/ /_/ / / / /_/ / __/ / / /_/ / |/ |/ /
/_/ |_/\___/\__,_/_/ \____/_/ /_/\____/|__/|__/
1.5.7
Network : neuroflow.nets.cpu.DenseNetwork
Weights : 2.480 (≈ 0,0189209 MB)
Precision : Double
Loss : neuroflow.core.SoftmaxLogEntropy
Update : neuroflow.core.Vanilla
Layout : 20 Vector
40 Dense (φ)
40 Dense (φ)
2 Dense (σ)
O O
O O
O O O
O O O
O O O O
O O O O
O O O
O O O
O O
O O
[run-main-0] INFO neuroflow.nets.cpu.DenseNetworkDouble - [09.03.2018 11:24:02:567] Training with 199 samples, batch size = 199, batches = 1.
[run-main-0] INFO neuroflow.nets.cpu.DenseNetworkDouble - [09.03.2018 11:24:02:635] Breeding batches ...
Mär 09, 2018 11:24:02 AM com.github.fommil.jni.JniLoader liberalLoad
INFORMATION: successfully loaded /var/folders/t_/plj660gn6ps0546vj6xtx92m0000gn/T/jniloader5703846428977288045netlib-native_system-osx-x86_64.jnilib
[run-main-0] INFO neuroflow.nets.cpu.DenseNetworkDouble - [09.03.2018 11:24:02:868] Iteration 1.1, Avg. Loss = 78,3524, Vector: 121.89397416123774 34.81072677614606
[run-main-0] INFO neuroflow.nets.cpu.DenseNetworkDouble - [09.03.2018 11:24:02:881] Iteration 2.1, Avg. Loss = 78,2962, Vector: 121.70090635111697 34.891518522053396
[run-main-0] INFO neuroflow.nets.cpu.DenseNetworkDouble - [09.03.2018 11:24:02:888] Iteration 3.1, Avg. Loss = 78,2376, Vector: 121.49919197504813 34.97606651932853
...
val cars = normalize(readAll("file/newsgroup/cars/", offset = maxSamples, max = maxSamples))
val med = normalize(readAll("file/newsgroup/med/", offset = maxSamples, max = maxSamples))
val testCars = vectorize(cars)
val testMed = vectorize(med)
def eval(id: String, maxIndex: Int, xs: Seq[Seq[Double]]) = {
val (ok, fail) = xs.map(net.evaluate).map(k => k.indexOf(k.max) == maxIndex).partition(l => l)
println(s"Correctly classified $id: ${ok.size.toDouble / (ok.size.toDouble + fail.size.toDouble) * 100.0} % !")
}
eval("cars", 0, testCars)
eval("med", 1, testMed)
Correctly classified cars: 98.98 % !
Correctly classified med: 97.0 % !
val free = normalize(readSingle("file/newsgroup/free.txt"))
val testFree = vectorize(free)
testFree.map(net.evaluate).foreach(k => println(s"Free classified as: ${if (k.indexOf(k.max) == 0) "cars" else "med"}"))
"They say there’s no such thing as a free lunch, and for doctors fed by drug companies, the old adage might be true. Even cheap meals provided by pharmaceutical sales reps were associated with higher prescription rates for the brands being promoted, a new JAMA Internal Medicine study concluded.
Researchers delved into data from the U.S. Open Payments database, mandated by the Sunshine Act, and matched it with Medicare Part D prescribing information on more than 276,000 doctors. They focused specifically on meals, rather than speaking fees or research payments.
They also focused on four on-patent drugs from four different drug classes, three treating cardiovascular issues and one antidepressant. AstraZeneca’s Crestor represented statin meds; Forest Laboratories’ Bystolic represented beta blockers; Daiichi Sankyo’s Benicar, ACE inhibitors and angiotensin receptor blockers; and Pfizer’s Pristiq, SSRI and SNRI antidepressants."
=> Free classified as: med
Source: http://www.fiercepharma.com/pharma/hey-doc-about-about-a-burger-a-side-branded-drug-scripts
"For the budget-conscious Tesla fans who can't wait for a Model 3, a cheaper version of the Model S is now available. Tesla said it has released a new Model S 60 Thursday which starts at $66,000. An all-wheel drive version starts at $71,000.The '60' in the Model S name refers to the battery size, which determines the range of the car on a single charge.
The new Model S 60 is estimated to go 210 miles on a single charge, which is a bit less than the range of the previous cheapest Model S. A slightly larger battery will let the '60' go 249 miles, but it'll cost you at least $74,500.
Even the $66,000 starting price still puts it firmly in luxury car territory, even though it is about $23,500 cheaper than the next cheapest version of the Model S -- the longer-range, all wheel drive Model S 90D that starts at $89,500.
The Model X, the Tesla crossover which debuted last year, has an $83,000 starting price."
=> Free classified as: cars
Source: http://money.cnn.com/2016/06/10/autos/cheaper-tesla-model-s/index.html